2022-scRNAseq数据可用的分析工具综述

| 分析类型 Analysis Type |
程序 Program |
数据集 Datasets |
|---|---|---|
| 细胞类型识别 Cell Identity |
SingleR (100) dynamicTreeCut (101) scHCL (102) CIBERSORT (bulk RNA seq) (103) |
HCL (102) |
| 基因腹肌分析 Gene Ontology |
EnrichR (104, 105) Metascape (107) clusterProfiler (108) GSVA (109) WebGestalt (110) |
MSigDB (106) Hallmark (111) REACTOME (112) KEGG HAdb (38) |
| 细胞周期状态 Cell State |
SCENIC/ (113) CellCycleScoring (97) |
|
| 细胞内互作 Intercellular Interaction |
FANTOM5 (114) STRING (115) CellPhoneDB (116, 117) CellChat (118) |
|
| 发育轨迹分析 Trajectory |
Monocle2/3 (119) Slingshot (120) SCORPIUS (121) Markov HC (122) RNA velocity (123) |
|
| 染色体变异 Chromosomal Variation |
inferCNV (44, 124) LIAYSON (55) VarTrix (23) GISTIC2 (125) Mutect2 (126) CopyKAT (127) |
|
| 生存分析 (Survival) |
Survminer (29) KaplanMeier Plotter (128) |
|
| 癌症基因表达 Cancer Gene Expression |
CIPHER (129) CancerSEA (130) GEPIA2 (131) UCSC Xena (132) |
TCGA STAD (133) Firebrowse (134) PanCanAtlas |
SCANPY是一个可用于scRNAseq数据预处理、聚类和差异基因表达分析的软件。 (99)
单细胞分析参考文献:Hoft, S. G., Pherson, M. D., & DiPaolo, R. J. (2022). Discovering Immune-Mediated Mechanisms of Gastric Carcinogenesis Through Single-Cell RNA Sequencing. Frontiers in immunology, 13, 902017. https://doi.org/10.3389/fimmu.2022.902017
2019-单细胞RNA序列技术和数据分析
scRNA-seq数据的各种分析概述
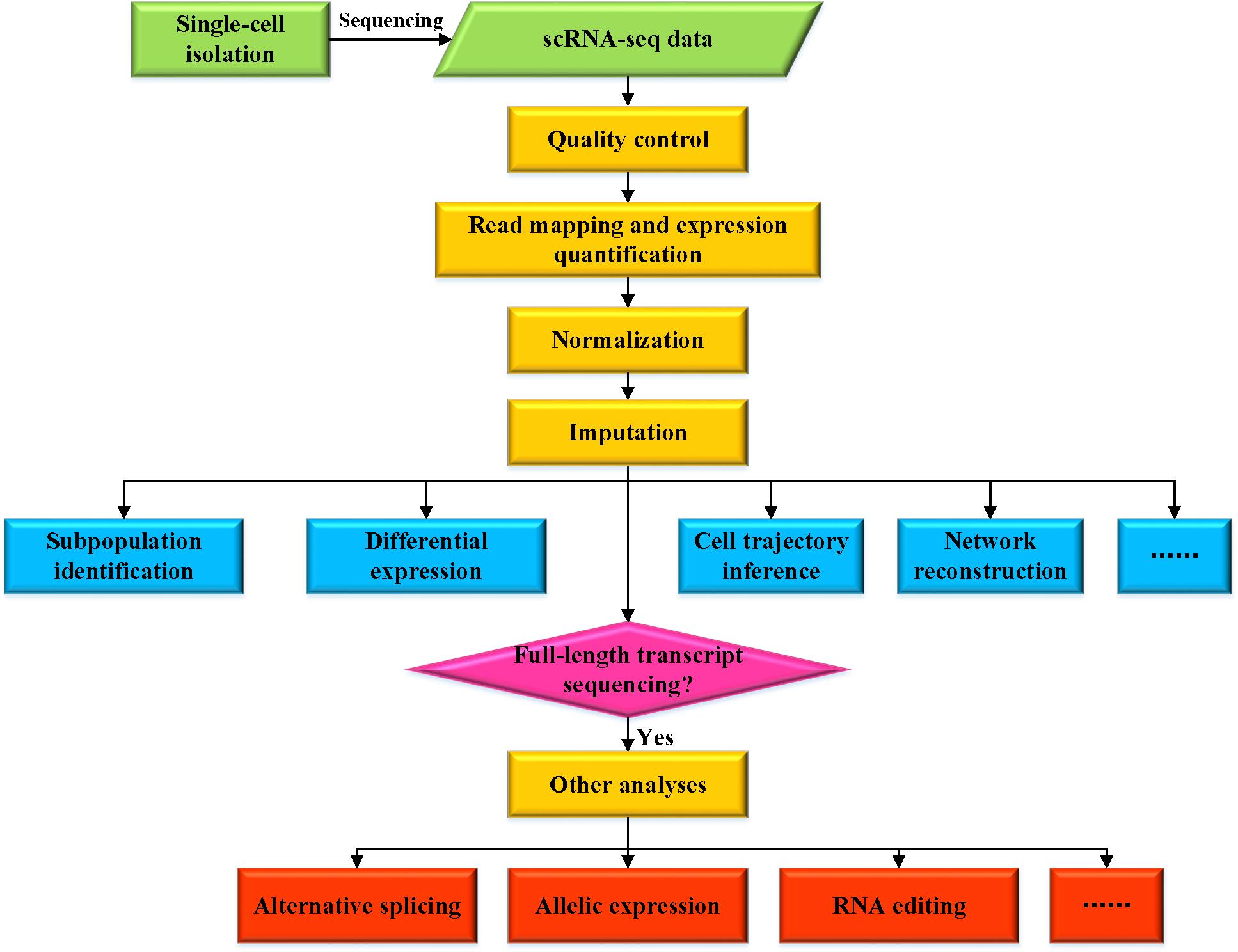
表1: scRNAseq测序技术
| Methods | Transcript coverage | UMI possibility | Strand specific | References |
|---|---|---|---|---|
| Tang method | Nearly full-length | No | No | Tang et al., 2009 |
| Quartz-Seq | Full-length | No | No | Sasagawa et al., 2013 |
| SUPeR-seq | Full-length | No | No | Fan X. et al., 2015 |
| Smart-seq | Full-length | No | No | Ramskold et al., 2012 |
| Smart-seq2 | Full-length | No | No | Picelli et al., 2013 |
| MATQ-seq | Full-length | Yes | Yes | Sheng et al., 2017 |
| STRT-seq and STRT/C1 | 5′-only | Yes | Yes | Islam et al., 2011, 2012 |
| CEL-seq | 3′-only | Yes | Yes | Hashimshony et al., 2012 |
| CEL-seq2 | 3′-only | Yes | Yes | Hashimshony et al., 2016 |
| MARS-seq | 3′-only | Yes | Yes | Jaitin et al., 2014 |
| CytoSeq | 3′-only | Yes | Yes | Fan H.C. et al., 2015 |
| Drop-seq | 3′-only | Yes | Yes | Macosko et al., 2015 |
| InDrop | 3′-only | Yes | Yes | Klein et al., 2015 |
| Chromium | 3′-only | Yes | Yes | Zheng et al., 2017 |
| SPLiT-seq | 3′-only | Yes | Yes | Rosenberg et al., 2018 |
| sci-RNA-seq | 3′-only | Yes | Yes | Cao et al., 2017 |
| Seq-Well | 3′-only | Yes | Yes | Gierahn et al., 2017 |
| DroNC-seq | 3′-only | Yes | Yes | Habib et al., 2017 |
| Quartz-Seq2 | 3′-only | Yes | Yes | Sasagawa et al., 2018 |
表2: 用于scRNA-seq数据比对和表达量计算的工具
| 工具 | 分类 | URL | 参考文献 |
|---|---|---|---|
| TopHat2 | Read mapping | https://ccb.jhu.edu/software/tophat/index.shtml | Kim et al., 2013 |
| STAR | Read mapping | https://github.com/alexdobin/STAR | Dobin and Gingeras, 2015 |
| HISAT2 | Read mapping | https://ccb.jhu.edu/software/hisat2/index.shtml | Kim et al., 2015 |
| Cufflinks | Expression quantification | https://github.com/cole-trapnell-lab/cufflinks | Trapnell et al., 2010 |
| RSEM | Expression quantification | https://github.com/deweylab/RSEM | Li and Dewey, 2011 |
| StringTie | Expression quantification | https://github.com/gpertea/stringtie | Pertea et al., 2015 |
表3:scRNA-seq数据的亚种群识别方法
| Methods | URL | References |
|---|---|---|
| SC3 | http://bioconductor.org/packages/SC3 | Kiselev et al., 2017 |
| ZIFA | https://github.com/epierson9/ZIFA | Pierson and Yau, 2015 |
| Destiny | https://github.com/theislab/destiny | Angerer et al., 2016 |
| SNN-Cliq | http://bioinfo.uncc.edu/SNNCliq/ | Xu and Su, 2015 |
| RaceID | https://github.com/dgrun/RaceID | Grun et al., 2015 |
| SCUBA | https://github.com/gcyuan/SCUBA | Marco et al., 2014 |
| BackSPIN | https://github.com/linnarsson-lab/BackSPIN | Zeisel et al., 2015 |
| PAGODA | http://hms-dbmi.github.io/scde/ | Fan et al., 2016 |
| CIDR | https://github.com/VCCRI/CIDR | Lin et al., 2017 |
| pcaReduce | https://github.com/JustinaZ/pcaReduce | Zurauskiene and Yau, 2016 |
| Seurat | https://github.com/satijalab/seurat | Satija et al., 2015 |
| TSCAN | https://github.com/zji90/TSCAN | Ji and Ji, 2016 |
表4:差异分析工具
| Methods | Category | URL | Referenes |
|---|---|---|---|
| ROTS | Single cell | https://bioconductor.org/packages/release/bioc/html/ROTS.html | Seyednasrollah et al., 2016 |
| MAST | Single cell | https://github.com/RGLab/MAST | Finak et al., 2015 |
| BCseq | Single cell | https://bioconductor.org/packages/devel/bioc/html/bcSeq.html | Chen and Zheng, 2018 |
| SCDE | Single cell | http://hms-dbmi.github.io/scde/ | Kharchenko et al., 2014 |
| DEsingle | Single cell | https://bioconductor.org/packages/DEsingle | Miao et al., 2018 |
| Cencus | Single cell | http://cole-trapnell-lab.github.io/monocle-release/ | Qiu et al., 2017 |
| D3E | Single cell | https://github.com/hemberg-lab/D3E | Delmans and Hemberg, 2016 |
| BPSC | Single cell | https://github.com/nghiavtr/BPSC | Vu et al., 2016 |
| DESeq2 | Bulk | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | Love et al., 2014 |
| edgeR | Bulk | https://bioconductor.org/packages/release/bioc/html/edgeR.html | Robinson et al., 2010 |
| Limma | Bulk | http://bioconductor.org/packages/release/bioc/html/limma.html | Ritchie et al., 2015 |
| Ballgown | Bulk | http://www.bioconductor.org/packages/release/bioc/html/ballgown.html | Frazee et al., 2015 |
表5:单细胞发育轨迹推断
| Tools | Dimensionality reduction | URL | 参考文献 |
|---|---|---|---|
| Monocle | ICA | http://cole-trapnell-lab.github.io/monocle-release/ | Trapnell et al., 2014 |
| Waterfall | PCA | https://www.cell.com/cms/10.1016/j.stem.2015.07.013/attachment/3e966901-034f-418a-a439-996c50292a11/mmc9.zip | Shin et al., 2015 |
| Wishbone | Diffusion maps | https://github.com/ManuSetty/wishbone | Setty et al., 2016 |
| GrandPrix | Gaussian Process Latent Variable Model | https://github.com/ManchesterBioinference/GrandPrix | Ahmed et al., 2019 |
| SCUBA | t-SNE | https://github.com/gcyuan/SCUBA | Marco et al., 2014 |
| DPT | Diffusion maps | https://media.nature.com/original/nature-assets/nmeth/journal/v13/n10/extref/nmeth.3971-S3.zip | Haghverdi et al., 2016 |
| TSCAN | PCA | https://github.com/zji90/TSCAN | Ji and Ji, 2016 |
| Monocle2 | RGE | http://cole-trapnell-lab.github.io/monocle-release/ | Qiu et al., 2017 |
| Slingshot | Any | https://github.com/kstreet13/slingshot | Street et al., 2018 |
| CellRouter | Any | https://github.com/edroaldo/cellrouter | Lummertz da Rocha et al., 2018 |
表6:scRNA-seq数据的可变剪接检测工具
| 工具 | URL | 参考文献 |
|---|---|---|
| SingleSplice | https://github.com/jw156605/SingleSplice | Welch et al., 2016 |
| Expedition | https://github.com/YeoLab/Expedition | Song et al., 2017 |
| BRIE | https://github.com/huangyh09/brie | Huang and Sanguinetti, 2017 |
| Census | http://cole-trapnell-lab.github.io/monocle-release/ | Qiu et al., 2017 |
Chen, G., Ning, B., & Shi, T. (2019). Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Frontiers in genetics, 10, 317. https://doi.org/10.3389/fgene.2019.00317